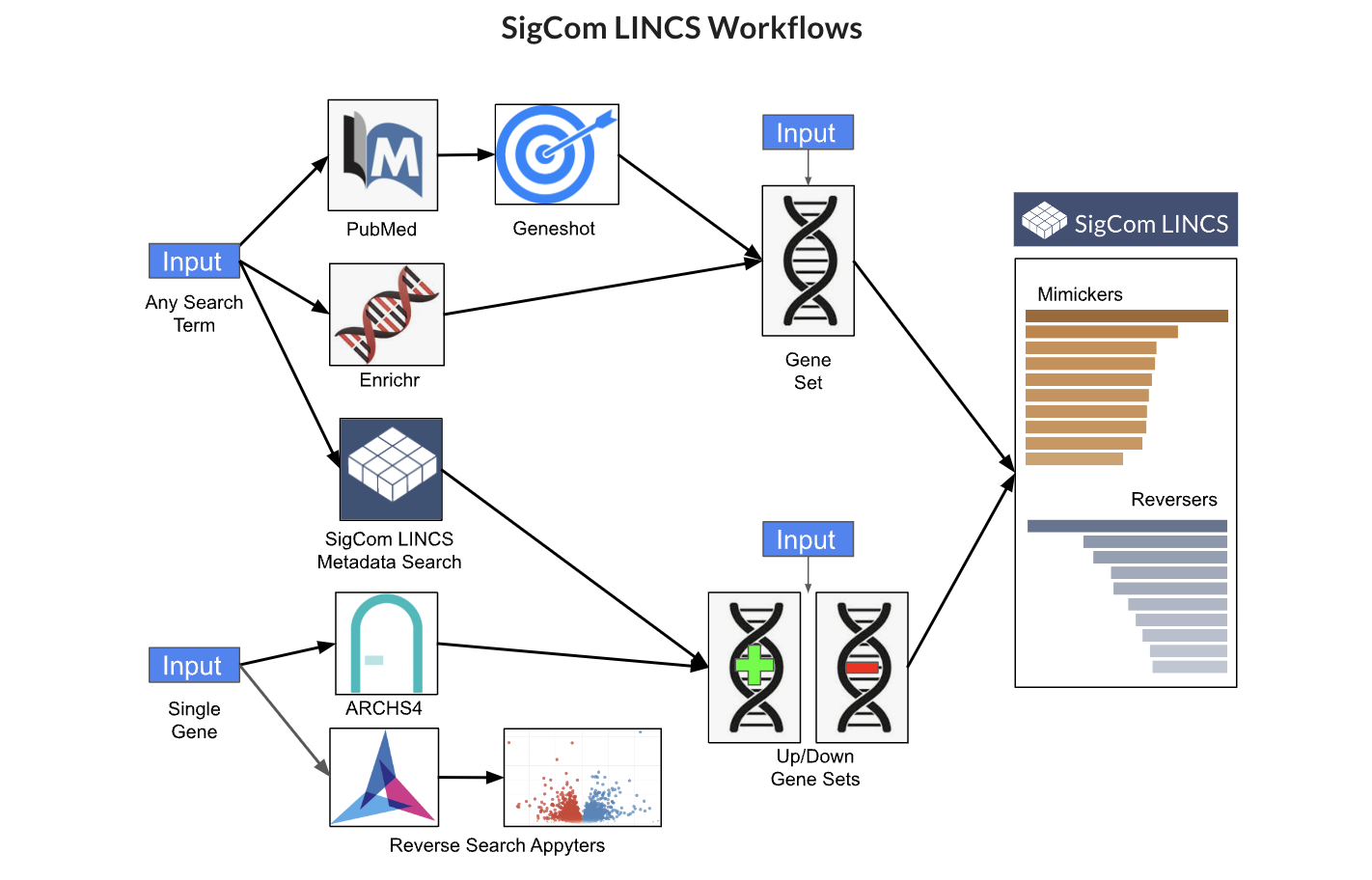
Enter a set of human gene symbols, or upload a file with human gene symbols, to the input form to receive an output from the various tools and databases.
In which annotated gene sets is my gene set enriched?
File formats accepted: csv, tsv, txt file with Entrez gene symbols on each line
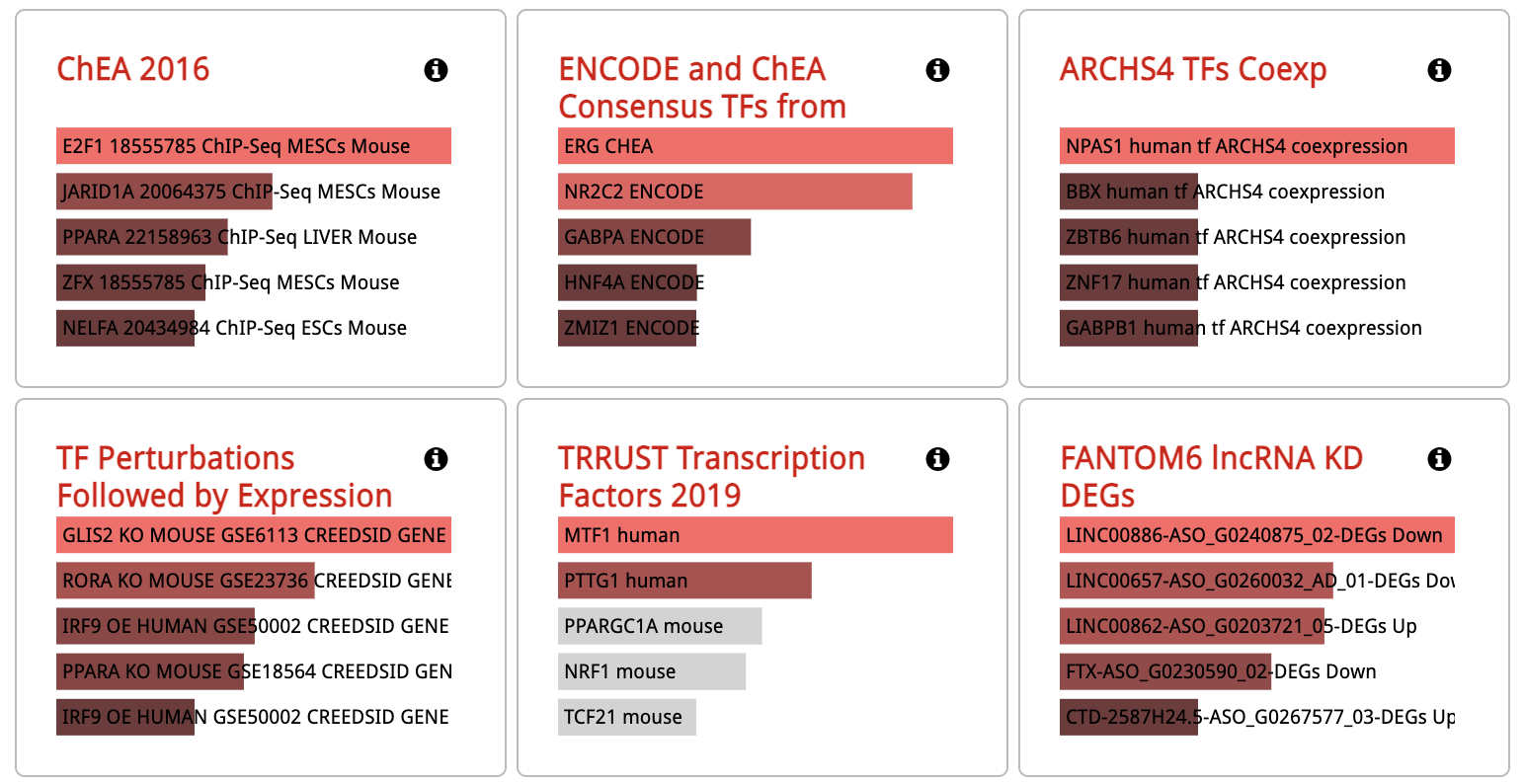
Perform enrichement analysis through Enricher with the entered or uploaded gene set:
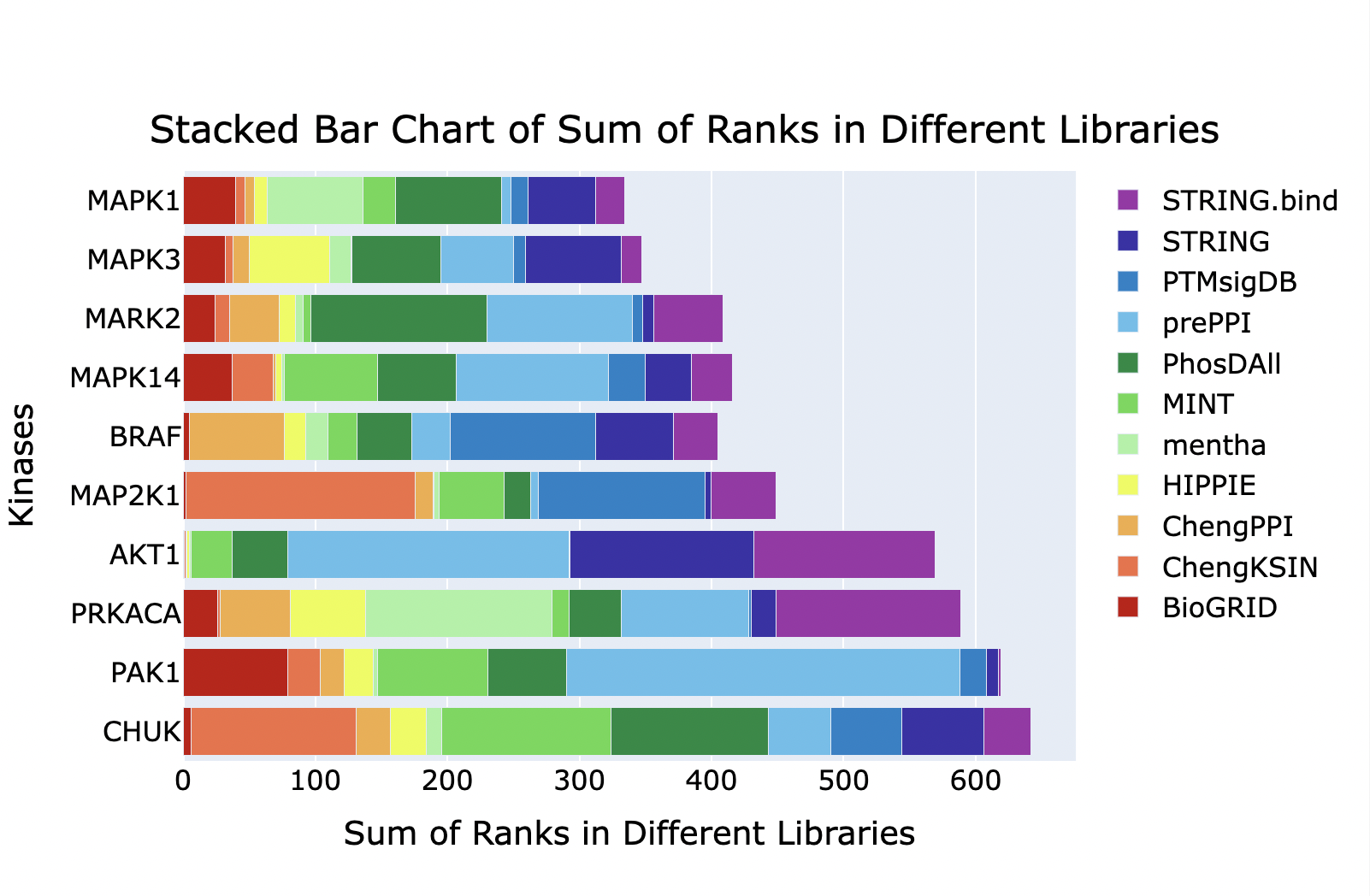
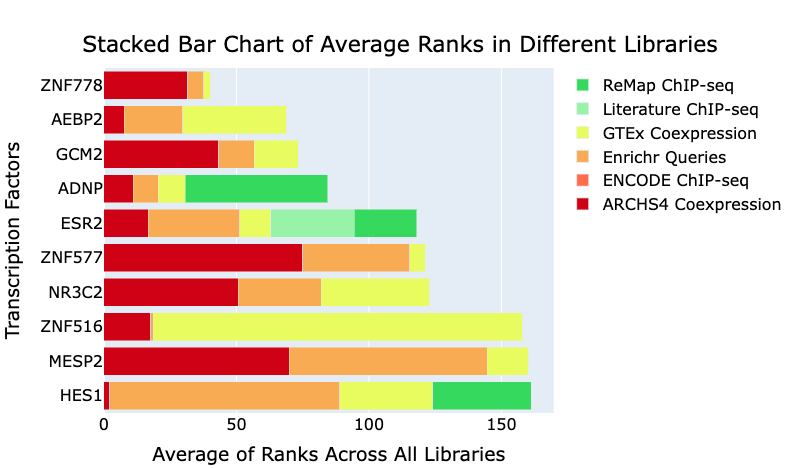
What transcription factors and kinases regulate my gene set?
File formats accepted: csv, tsv, txt file with Entrez gene symbols on each line
The ChEA3 Appyter (ChIP-X Enrichment Analysis 3) predicts transcription factors (TFs) associated with user-input sets of genes. Discrete query gene sets are compared to ChEA3 libraries of \TF target gene sets assembled from multiple orthogonal 'omics' datasets.
What are the LINCS L1000 small molecules and genetic perturbations that likely up- or down-regulate the expression of my gene set?
File formats accepted: csv, tsv, txt file with Entrez gene symbols on each line
Query SigCom LINCS which is a web-based search engine that serves over 1.5 million gene expression signatures processed, analyzed, and visualized from LINCS, GTEx, and GEO. SigCom LINCS provides rapid signature similarity search for mimickers and reversers given sets of up and down genes.